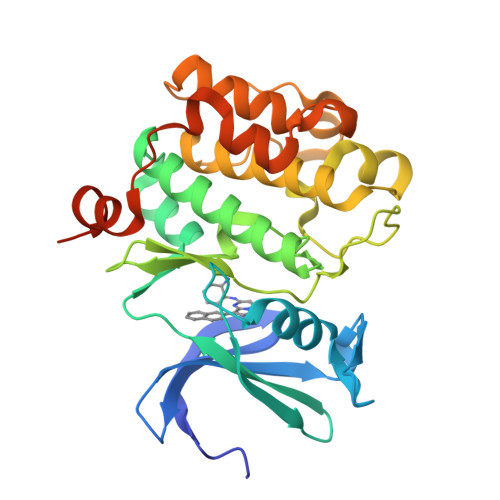
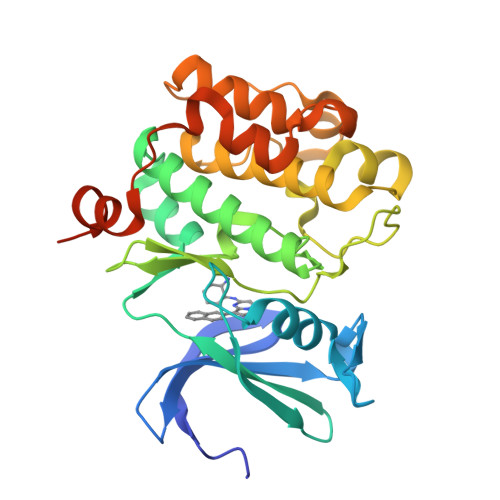
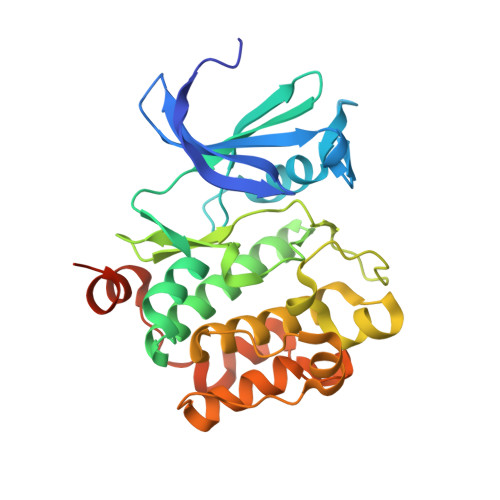
Discovery of pyrazolo[1,5-a]pyrimidine-based Pim inhibitors: A template-based approach.
Dwyer, M.P., Keertikar, K., Paruch, K., Alvarez, C., Labroli, M., Poker, C., Fischmann, T.O., Mayer-Ezell, R., Bond, R., Wang, Y., Azevedo, R., Guzi, T.J.(2013) Bioorg Med Chem Lett 23: 6178-6182
- PubMed: 24091081
- DOI: https://doi.org/10.1016/j.bmcl.2013.08.110
- Primary Citation of Related Structures:
4MBI, 4MBL - PubMed Abstract:
The synthesis and hit-to-lead SAR development from a pyrazolo[1,5-a]pyrimidine-derived hit 5 to the identification of a series of potent, pan-Pim inhibitors such as 11j are described.
Organizational Affiliation:
Department of Chemical Research, Merck Research Laboratories, 2015 Galloping Hill Road, Kenilworth, NJ 07033, United States. Electronic address: michael.dwyer@merck.com.